SteinBlog
nmrML: A vendor-neutral open exchange format for NMR-based metabolomics
Metabolomics is a growing field and the number of organisms being studied is constantly increasing as is the number of metabolites being discovered. With this growth comes a steady increase in the amount of metabolomics data and the need to ensure that we capture this information in persistent open formats and databases.
The COSMOS project (COordination Of Standards In MetabOlomicS, http://www.cosmos-fp7.eu/) has been created to improve the adoption of open standards for metabolomics data, annotation with agreed metadata, and support by open source data management and capturing tools. COSMOS delivers an ecosystem of formats, tools, and resources such as MetaboLights (http://www.ebi.ac.uk/metabolights/), a database for capturing information obtained in metabolomics experiments.
In a spotlight in the December 2014 issue of MetaboNews we introduce recent developments in nmrML (www.nmrml.org), a vendor-neutral open exchange and storage format to describe NMR-based metabolomics data.
Structure Elucidation of Unknown Metabolites (2) – De Novo vs Look-Up
There has always been a bit of confusion in the terminology around the subject of computer-assisted structure elucidation (CASE), so let’s define some terms:
- Structure Elucidation – Determining the structure of truly unknown compounds de-novo from spectroscopic (or earlier by hard core chemistry :)) experiments
- Structure Identification – Recovering the structure of already known compounds from databases or printed reference material (including experimental sections of the primary literature)
- Dereplication: Same as 2.
The more we discover, the more likely it will be that we are able to de-replicate by database lookup. This of course requires well curated and developed open access databases that cover many chemical compounds/metabolites.
In organic chemistry, spectroscopic databases for structure identification where published quite early puttygen download , albeit as closed-access, commercial systems. The most widely used examples is probably the SpecInfo database which now seems to be marketed by Wiley and the more recently (considering the 40-year horizon of the topic :)) published ACD/Labs spectral libaries and management system. Wolfgang Robien in Vienna has been developing NMR spectral databases and prediction tools for a long time.
The general way of searching in such databases would be to measure an NMR spectrum of your isolated unknown compound, perform a peak picking and search the database using this peak picking (a feature vector, if you wish).
In the early 2000’s my Stephan Kuhn in my group developed the NMRShiftDB database which was the first open access, open source, open submission, web-based NMR database where you can now test how this all works without running into pay walls. Stephan has left the lab and now runs version 2 of this database in collaboration with the NMR lab at the department of chemistry at the University of Cologne.
One caveat: It is much easier to search for carbon-13 NMR spectra or mass spectra than for proton NMR spectra. The latter has rarely been addressed, not the least because of the lack of full spectrum proton data to which you could match a real-life proton spectrum. Peak-picking proton NMR spectra is problematic often due to overlap and complex coupling patterns.
Take for example the carbon-13 spectrum of pinocarveol, both from the metabolomics section of BioMagResBank (BMRB). Using your NMR software’s peak picking method, you would end up with this list of NMR signals. If you have a decent browser, such as FireFox, you can use the CMD (on mac) or CTRL key and select the chemical shifts in the table linked above. If not, here they are:
158.786
114.081
69.704
53.211
43.079
42.498
37.114
30.571
28.588
28.588
24.609
24.609
Structure Elucidation of Unknown Metabolites (1) – Problem Description
In preparation for a talk about structure elucidation of unknown chemical compounds at AISBM in Paris in October, I laying out a bit of the work that others as well as my group have done over the past 40 years in this area. The topic of this AISBM meeting is “Challenges and advances in the annotation and de novo identification of small molecules of biological origin”. I am going to address the problem of computer-assisted structure elucidation (CASE) of unknown compounds in organic chemistry in general and refer to the sub-problem of natural products and metabolites when needed.
Generally speaking, we are talking about the problem where you have an evidence that there is a compound in a biological system or your flask but you don’t know the structure of it. By structure, I mean ideally the fully defined stereo isomer but at least the fully defined constitutional isomer.
I have reviewed the problem of computer-assisted structure elucidation (CASE) a few times in the context of Natural Products Structure Elucidation (Steinbeck 2001, Steinbeck 2004).
Assume, for example, that you are working on a newly discovered medicinal plant and want to discover the compound or set of compounds responsible for the activity.

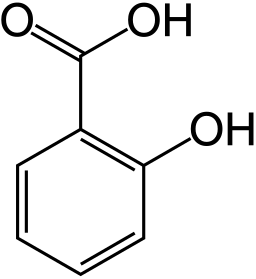
Willow Tree (Courtesy of Wikipedia). The bark contains Salicylic Acid and was used for a long time as a pain reliever and fever reliever.
How could one find out that the structure of the active ingredient is as follows?
The evidence you’ve got could come from a chromatographic experiment where you have become interested in a particular peak that shows a biological activity.

HPLC chromatogram of a perfume mixture (courtesy of Wikipedia). Each of those peaks is at least one compound. What is the structure of the compound under the leftmost signal?
In our context, the information for determining this information comes from spectroscopic information – NMR and/or Mass Spectrometry (MS). In order to retrieve this information, we need to isolate the compound using separation techniques such as HPLC or use hyphenation techniques.
In the next post, I will elaborate on some of case scenarios that we might be facing in structure elucidation.
References:
Steinbeck, C. “The Automation of Natural Product Structure Elucidation..” Current Opinion in Drug Discovery and Development 4.3 (2001): 338–342. Print.
Steinbeck, C. “Recent Developments in Automated Structure Elucidation of Natural Products.” Natural Product Reports 21.4 (2004): 512–518. Print.
Journal of Cheminformatics Impact Factor gone up to 4.54
I am delighted to report that the latest Impact Factor for the Journal of Cheminformatics is 4.54 which is a huge improvement over the 3.59 from 2013. Congratulations and thanks to out authors PuTTY , the editorial and production team, our editorial board and my co-editor-in-chief David Wild for making this possible.
DIY Bookscanner Assembly
I have written about the arrival and packaging of my bookscanner kit from diybookscanner.eu in a previous post.
Here I document the assembly, which was really easy due to the excellent videos by Daniel Reetz on YouTube. I recommend watching them all before even starting to do anything with the kit. Regarding tools needed for the assembly: An electric drill and an electric screw driver are really handy. You’ll need a Philips and a hex tip screwdriver. A rubber hammer is useful but a normal hammer and a piece of scrap wood will do. I am not going to repeat what Daniel has documented so perfectly but just show a series of pictures that document the assembly. This is essentially done in an afternoon.

https://lookup-phone-prefix.com
, which will hold the glass and the cameras” width=”300″ height=”225″ /> The Imaging Module, which will hold the glass and the cameras
DIY Bookscanner Kit arrived
I love books. Real books, made of paper, ideally hard cover and properly bound with needle and thread. And I have assembled a nice library over time.
On the other hand, due to peculiar choices of where to live and where to work, and due to the nature of my job, I essentially live on the road and in the air. That is incompatible with carrying around heavy, beautiful and sensitive books.
Over the years I have more and more reduced the weight of my backpack and recent improvements in offline reading apps for the magazines I read (C’t, Make Magazine, Nature) make it possible that I can now read all of them in the air and offline on my iPad without carrying around the paper and its weight. At any given time, however, I am also reading a book from my tall FIFO stack of books. Buying them as ebooks is not an option for me, due to my love for physical books and other concerns about DRM and loss of access.
Due to another passion, Makeing, Open Source Software and Open Hardware, I came across the fantastic DIYBookScanner project. After Daniel Reetz, a key figure of this movement, released the plans for an open hardware standard bookscanner kit, I knew it was time. Because of time constraints, I decided to buy pre-cut parts from http://diybookscanner.eu/. I hope to build one from scratch soon at MakeSpace Cambridge.
But for now I would like to report on my progress with the book scanner kit from http://diybookscanner.eu. I decided to buy the base kit and get the cameras, foot pedal and USB hub from Amazon.
The kit arrived in a nice, light and compact package. The purchase includes video chat support for the assembly, which I didn’t need due to the extensive online documentation.
Parts where each wrapped in shrink wrap.
I will report on putting this together in a following post. For now I would like to congratulate the folks at http://diybookscanner.eu/ for their excellent product.
Bioinformatics PhD opening for UK national
Towards a better understanding of lipid metabolism through studies of Drosophila Lipidomics
Christoph Steinbeck, Julian Griffin, Steve Russell
This project will bring together Drosophila experts at the University of Cambridge (Prof. Steve Russell, Dept of Genetics and Cambridge Systems Biology Centre), the Lipidomics lab led by Julian Griffin at the MRC Unit for Human Nutrition Research/Biochemistry, University of Cambridge and the Computational Metabolomics Group led by Christoph Steinbeck at the European Bioinformatics Institute in Hinxton to extensively study the lipidomics of Drosophila. The student will grow up a number of Drosophila mutant models related to lipid metabolism, isolate fat pads and perform analysis using high resolution mass spectrometry. The data will be modelled within the Computational Metabolomics Group to identify differences between the mutants, and explore how the Drosophila lipidome is regulated.
For this BBSRC-funded project, only candidates with a UK passport are eligible. Please email steinbeck@ebi.ac.uk if you are interested.
In Memory of Open Science Pioneer Jean-Claude Bradley

Jean-Claude Bradley receiving the Blue Obelisk award presented by Egon Willighagen in an improvised ceremony during the ACS meeting in Chicago 2007 (Chris Steinbeck behind the camera). Picture licensed CC-BY.
As announced today by Drexel University, Open Science Pioneer and recipient of the Blue Obelisk award Jean-Claude Bradley died yesterday. He was a member of the editorial board of the Journal of Cheminformatic and Editor-in-Chief of the Chemistry Central Journal. But most importantly, he was an inspiring scientist and evangelist of the open science and open access movement.
Open PhD position in Steinbeck group (Cheminformatics and Metabolism)

Where we live: EBI Main Building on Genome Campus
We have an opening for a Ph.D. position in Chris Steinbeck’s cheminformatics and metabolism team at the European Bioinformatics Institute (EBI) in Cambridge, UK.
Ph.D. topics are available in a wide range of areas such as analysis of metabolomics experiments, metabolism, computational natural product biochemistry, elucidation of natural products, text mining and image processing, and more. Applications should be submitted through the EMBL International Ph.D. programme. Ph.D. students successfully pursuing and completing their projects will retrieve their Ph.D. from the University of Cambridge, UK.
The group leaders at the EBI recruiting in this round are Sarah Teichman, Alex Bateman, John Overington and Chris Steinbeck. Candidates invited to the final assessment have the opportunity to consider all of the recruiting groups. The registration and submission deadlines are approaching quickly.